Comparative Analysis of Yukmijihwang-tang and Palmijihwang-tang: Lifespan Extension and Kidney Function Improvement in Aging Models
Ⓒ The Society of Pathology in Korean Medicine, The Physiological Society of Korean Medicine
Abstract
Aging is associated with a decline in physical and mental functions, necessitating support strategies to extend the healthy life expectancy (HLE). Traditional Asian herbal medicines, such as Yukmijihwang-tang (YM) and Palmijihwang-tang (PM), are used to enhance HLE by improving the kidney function and treating age-related conditions. In this study, we aimed to investigate the effects of YM and PM on lifespan extension and kidney aging marker and aging-related gene expression levels in mice and elucidate the underlying hormonal mechanisms. A lifespan assay was conducted using Caenorhabditis elegans to compare and analyze the effects of YM and PM on HLE and kidney function. Subsequently, aging markers and kidney function were analyzed in aged mice treated with YM and PM. Additionally, RNA sequencing of kidneys was performed to analyze the RNA expression levels related to the hormonal pathways of YM and PM. YM and PM significantly increased the lifespan of C. elegans in a dose-dependent manner. In aged mice, YM and PM restored the kidney aging markers, including the plasma klotho and albumin levels, glomerular filtration rate, urine volume, osmolality, and sodium levels. Transcriptomic analysis revealed distinct gene expression patterns. Although both YM and PM regulated similar pathways, PM affected 4.5-times more genes associated with the mitochondria, endoplasmic reticulum, and their membranes than YM. Furthermore, YM and PM modulated the hormone metabolism genes associated with longevity, highlighting their potential to mitigate age-related conditions through diverse molecular pathways. This study demonstrated the potential of YM and PM to extend HLE by modulating gene expression and improving kidney function, thus providing promising natural therapeutics with few side effects.
Keywords:
Aging, Yukmijihwang-tang, Palmijihwang-tang, Healthy life expectancy, KidneyIntroduction
Aging is a gradual biological process characterized by the irreversible decline in physical and mental functions, including the accumulation of cellular and tissue damage, loss of homeostasis, increased risk of various aging-related diseases, and high mortality rate1,2). The elderly population is rapidly expanding worldwide and is expected to surpass two billion by 2050, constituting over 20% of the global population3). Along with the increase in the elderly population, average life expectancy also increased until the coronavirus disease-2019 pandemic4), leading to substantial interest in extending the healthy life expectancy (HLE), which differs from the biological lifespan, of individuals5). HLE is a metric that combines information on mortality and health status that is defined as the average number of years an individual is expected to live in a state of good health.
Many studies have explored methods to increase HLE, with key strategies including regular exercise, smoking cessation6), and adoption of a healthy diet7). Some studies have proposed metformin as a pharmacological agent to extend HLE, citing its benefits in improving metabolic health, reducing oxidative stress, and promoting cellular maintenance8). However, long-term use of metformin can lead to side effects, such as vitamin B-12 deficiency9), gastrointestinal disorders10), and lactic acidosis11), which limit its use.
In the East, traditional herbal medicines, such as Yukmijihwang-tang (YM) and Palmijihwang-tang (PM), which exert anti-aging effects and improve the kidney function, are used to enhance HLE. Both YM and PM share the same six herbs, but PM includes two more: Cinnamomi cortex (CC) and Aconiti lateralis (AL). These additions shift its focus from nourishing the Kidney Yin to supporting the Kidney Yang, changing its therapeutic use.
YM alleviates age-related conditions, such as weakness, memory decline, dizziness, geriatric diseases, and Parkinson’s disease, thereby increasing the HLE12-14). Palmijihwang-tang (PM) is used to treat age-related endocrine function decline, energy metabolism disorders, and ophthalmic symptoms, such as cataracts15,16).
However, these uses are inconsistent with the historical context in which YM and PM have been used to treat and prevent aging by improving the symptoms of kidney dysfunction through hormonal regulation17-19) as well as with the association between kidney function and longevity20). Historically, YM has been used to nourish the Kidney Yin, addressing symptoms such as dizziness, a spinning sensation in the eyes, ringing in the ears, night sweats, dry mouth and throat, and constipation21-23). In contrast, PM is formulated to support Kidney Yang, making it effective for symptoms such as low body temperature, cold hands, and feet, pale complexion, sore and aching knees, and issues like erectile dysfunction or infertility15).
The mechanisms by which YM affects aging to extend HLE must be analyzed, particularly focusing on the kidney function and hormonal pathways. Furthermore, effect of PM on HLE and its biological mechanisms should be investigated. These studies can aid in the development of new herbal medications to extend HLE with fewer side effects than metformin.
In this study, we aimed to verify the lifespan-extending effects of YM and PM using both Caenorhabditis elegans and mouse models. We analyzed the changes in kidney function and aging markers in aged mice after YM and PM treatments. Furthermore, we conducted RNA sequencing analysis of the kidneys of aged mice to compare the mechanisms by which YM and PM restored the age-related gene expression abnormalities. We also analyzed the hormonal pathways influenced by YM and PM by analyzing the hormone-related genes. This study offers potential benefits and novel insights, as it seeks to verify that YM and PM can enhance HLE and aims to reveal their mechanisms, thereby establishing a biological connection to the symptom clusters with which they have been historically associated.
Materials and Methods
1. Preparation and analyses of YM and PM
YM was extracted from the following medicinal herbs purchased from Omniherb (Daegu, Republic of Korea): Rehmannia glutinosa Liboschitz ex Steudel, Dioscorea japonica Thunberg, Cornus officinalis Siebold et Zuccarini, Paeonia suffruticosa Andrews, Poria cocos Wolf, and Alisma orientale Juzepzuk. PM was extracted from the same herbs, along with two other herbs, CC and AL. The herbal mixtures were refluxed twice in distilled water (10 L/kg) for 3 h, and the extracts were filtered, concentrated using a rotary evaporator, and dried using a freeze dryer. All herbarium voucher specimens (Nos. KYM2_2023 and KPM2_2023) and medicinal herbs were deposited at the Korea Institute of Oriental Medicine (KIOM, Daejeon, Korea).
The PM and YM extracts were dissolved in 50% methanol solution. The samples were analyzed via ultra-high-performance liquid chromatography (UHPLC; 1290 Infinity II LC system; Agilent Technologies, Santa Clara, CA, USA) combined with quadrupole time-of-flight mass spectrometry (Q-TOF MS; 6546 Q-TOF, Agilent Technologies) using the Zorbax Extend-C18 column (2.1 x 50 mm, 1.8 μm, Agilent Technologies) with a column temperature of 40℃. The solvent system consisted of water with 0.1% formic acid as solvent A and acetonitrile with 0.1% formic acid as solvent B at a flow rate of 0.1 mL/min. The solvent gradient was set as follows: 0–3 min, 95% A and 5% B; 3–5 min, 50% A and 50% B; 5–20 min, 15% A and 85% B; 20–23 min, 15% A and 85% B; 23–24 min, 85% A and 15% B; 24–30 min, 95% A and 5% B. Data acquisition and annotation were performed using MS-DIAL (version 4.90) and Agilent Mass Hunter (version 10.0).
2. Lifespan assay
As C. elegans survival assays are key methods to identify novel genetic factors regulating aging and lifespan in animals24), we used the wild-type N2 C. elegans model to investigate the potential effects of YM and PM on lifespan. The nematode was obtained from the Caenorhabditis Genetics Center at the University of Minnesota (Minneapolis, MN, USA) and cultured at 20℃ on the nematode growth medium (NGM) agar plates supplemented with Escherichia coli OP50, as previously described25).
Lifespan assays were conducted independently under standard culture conditions at least four times. Based on preliminary results indicating optimal lifespan extension effects among the five concentrations, 100 μg·mL−1 and 200 μg·mL−1 in C. elegans, the nematode species were synchronized using embryo isolation and L1 arrest techniques, and transferred to NGM plates at concentrations of 100 and 200 μg·mL−1, respectively to select the optimal dosage. After confirming 200 μg·mL−1 as the optimal dose, we further verified the effects of 200 µg·mL−1 YM and PM compared to those of the control.
Nematode species were also treated with two herbs present in PM (CC and AL), but not in YM, at concentrations of 10, 50, 100, and 200 μg·mL−1, and the results were compared with those of the control. Nematodes that did not respond to contact with the platinum wire tip were considered to be dead. To ensure consistent conditions, live nematodes were transferred to fresh NGM plates every 2 d. Survival rates were monitored using the SMZ1500 dissecting microscope (Nikon, Tokyo, Japan).
3. Animal study
All animal experiments adhered to the care and use guidelines for laboratory animals and ARRIVE 2.0 guidelines. This study was approved by the Institutional Animal Care and Use Committee of KIOM (approval number: 23-004).
Fourteen-month-old male C57BL/6 mice (n = 27) were obtained from Saeron Bio Co. (Uiwang, Republic of Korea) and maintained under specific pathogen-free conditions. After a five-month acclimatization period, the mice were divided into three groups: OLD (natural aging group), YM (100 mg/kg/day YM orally 5 d a week), and PM (same dosage and schedule as YM) groups. Additionally, young (Y) mice were included as a control group. Administration of YM and PM began when the mice were 19-month-old and continued until they were 24-month-old. Three-month-old C57BL/6 mice were used as young controls to compare with the old groups. These young mice underwent a one-month acclimatization period before the experiments. After the experiments, all mice were fasted for 12 h and blood and tissue samples were collected. Kidney tissues were promptly frozen at −80℃ for RNA sequencing analysis. Prior to analysis, random sampling was conducted owing to the limited samples. Furthermore, data points outside the measurement confidence interval were excluded from analysis.
4. Metabolic and renal function analyses
Urine samples were collected to determine the creatinine levels, sodium (Na+) levels, and osmolality using metabolic cages for 24 h. The collected urine samples were centrifuged at 2000 × g for 15 min at 4℃, and the supernatants were stored in a deep freezer until use. Ion concentrations were measured using the Electrolyte Analyzer (NOVA Biochemical, Waltham, MA, USA), and osmolality was measured using an Advanced Osmometer (Advanced Instruments Inc., Norwood, MA, USA). Creatinine concentrations in plasma and urine were measured using enzyme-linked immunosorbent assay (ELISA) kits (#ab285231; Abcam, Cambridge, MA, USA). Glomerular filtration rate (GFR) was estimated based on creatinine clearance (Ccr). Ccr was calculated as follows: Ccr = (Ucr × V) / Pcr, where Ucr is the urinary creatinine concentration, V is the volume of urine, and Pcr is the plasma creatinine level. Plasma klotho (Kl) levels were measured using Mouse Klotho ELISA kits (MOFI00553; Assay Genie, Ireland) and albumin levels were measured using Mouse Albumin ELISA kits (#ab207620; Abcam).
5. RNA isolation and sequencing
Total RNA was extracted from mouse kidney tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA quality was assessed using the Agilent TapeStation system (Agilent Technologies, Amstelveen, the Netherlands), and RNA quantification was performed using the Qubit fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Samples from each experimental group were randomly selected and analyzed by Ebiogen Inc. (Seoul, Republic of Korea).
QuantSeq 3'-mRNA-Seq Library Prep Kit (Lexogen, Inc., Vienna, Austria) was used to create RNA libraries for both the control and experimental samples. The sequencing reads from the kit were aligned via spliced transcript alignment to a reference26), and differential expression analysis was conducted using HTseq-count, a tool developed using HTSeq that pre-processes the RNA-seq data27), or both unique and multiple alignments. Read-count data were normalized using the trimmed mean of M values and counts per million mapped reads implemented in EdgeR.
Gene classification was conducted by searching the Database for Annotation, Visualization, and Integrated Discovery (http://david.abcc.ncifcrf.gov/) and Medline (http://www.ncbi.nlm.nih.gov/). Additional analyses were conducted on hormone-related genes using ClueGO, a Cytoscape plug-in that improves the biological interpretation of multiple genes. Data mining and visualization were performed using the Excel-based differentially expressed gene (DEG) analysis software (Ebiogen Inc.).
6. Statistical analyses
Data of lifespan experiments were illustrated using Kaplan–Meier survival curves, and differences in survival among groups were examined using the log-rank test. Data are expressed as the mean ± standard deviation. Comparisons between and among groups were performed using the Student’s t-test and one-way analysis of variance, followed by Tukey’s post-hoc test, respectively, with GraphPad Prism version 10 (GraphPad Software, La Jolla, CA, USA). Statistical significance was set as follows: *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
1. Chemical components of YM and PM
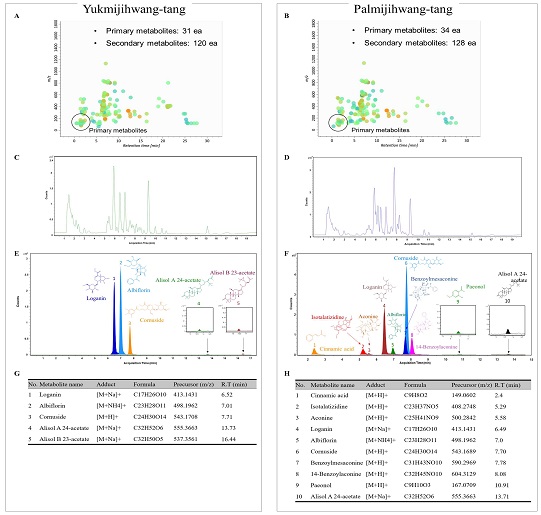
The major chemical constituents of YM and PM were confirmed to be compounds that met the criteria of relative retention time, precursor m/z, and MS/MS fragmentation patterns in UHPLC Q-TOF MS analysis. In total, 4647 (YM) and 4552 (PM) naked features were detected, and 151 metabolites of YM and 162 metabolites of PM were putatively matched with the reference peaks of the primary and secondary metabolites. We identified five major compounds in YM: loganin, albiflorin, cornuside, alisol A 24-acetate, and alisol B 23-acetate. In PM, we identified 10 major compounds: cinnamic acid, isotalatizidine, aconine, loganin, albiflorin, cornuside, benzoylmesaconine, 14-benzoylaconine, paeonol, and alisol A 24-acetate. Four major compounds were common in both YM and PM. The peaks of loganin and albiflorin were higher in YM than in PM, whereas those of cornuside and alisol A 24-acetate were higher in PM than in YM(Fig. 1).
2. YM and PM significantly increase the mean lifespan of C. elegans, thus exerting lifespan-extending effects
To determine the optimal dose, we treated C. elegans with YM and PM at concentrations of 100 and 200 μg·mL−1. The optimal dose was 200 μg·mL−1, resulting in mean lifespan of 21.40 (±0.81) d with YM and 22.55 (±0.86) d with PM (Fig. 2A and B). Subsequently, we verified the effects of YM and PM at 200 μg·mL−1. At this dose, the mean lifespan was 21.79 (±0.86) d with YM and 23.05 (±0.91) ds with PM compared to 17.68 (±0.69) d with the control. The maximum lifespan was 34 d with YM, 36 d with PM, and 29 d with the control. Lifespan increase was 23.21% with YM (p < 0.001) and 30.35% with PM (p < 0.001), with PM extending the lifespan by 7.14 percentage points compared to YM (Fig. 2C and D).
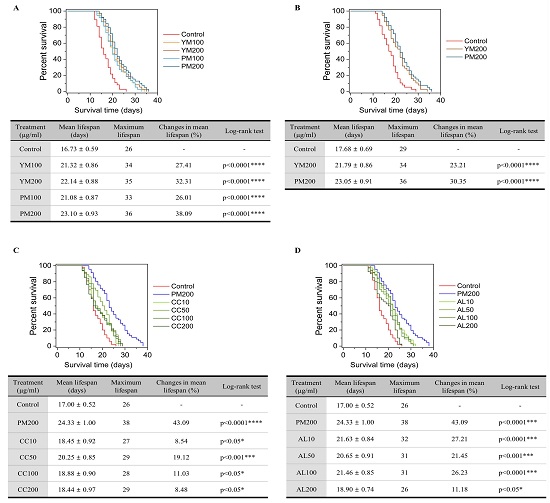
Effects of Yukmijihwang-tang (YM) and Palmijihwang-tang (PM) on the lifespan of wild-type N2 Caenorhabditis elegans. (A and B) Effects of YM and PM at 100 and 200 μg·mL−1. (C and D) Effects of YM and PM at 200 μg·mL−1. (E and F) Effects of PM at 200 μg·mL−1 and Cinnamomi cortex (CC) at 10, 50, 100, and 200 μg·mL−1. (G and H) Effects of PM at 200 μg·mL−1 and Aconiti lateralis (AL) at 10, 50, 100, and 200 μg·mL−1. All treatments were compared to the control group. Mean lifespan is represented as the mean ± standard error of the mean. Data for the control and 200 μg·mL−1 PM groups are the same in (E and F) and (G and H) and are shown separately for convenience to compare CC and AL, respectively.
As PM exhibited more potent lifespan-extending effects than YM, we clarified the components contributing to these effects by treating the C. elegans model with CC and AL, each at concentrations of 10, 50, 100, and 200 μg·mL−1. The highest mean lifespan was 20.25 (±0.85) days with 50 μg·mL−1 of CC (Fig. 2E and F) and 21.63 (±0.84) days with 10 μg·mL−1 of AL (Fig. 2G and H).
3. YM and PM restore the kidney aging markers in mice
YM and PM were administered to the mice starting at 19 months of age and continued until they reached 24 months, at which point they were sacrificed (Fig. 3A). Although no significant differences were observed in the survival rates of the OLD and herbal treatment groups (YM and PM) at the time of sacrifice, notable differences were observed in the kidney aging indicators. Plasma Kl levels were significantly lower in the OLD group (18.0 pg/mL) than in the Y group (49.5 pg/mL, p < 0.05) but significantly increased in the YM (45.6 pg/mL, p < 0.01) and PM (42.9 pg/mL, p < 0.05) groups (Fig. 3B). Plasma albumin levels were significantly higher in the OLD group (27.3 g/dL) than in the Y group (2.1 g/dL, p < 0.001) but significantly decreased in both the YM and PM groups (2.2 g/dL, p < 0.001; Fig. 3C). GFR was significantly decreased in the OLD group (60.6 mL/kg/h) compared to that in the Y group (348.7 mL/kg/h, p < 0.01) but increased in the YM (107.8 mL/kg/h) and PM (151.4 mL/kg/h) groups, without statistical significance (Fig. 3D). Urine volume was increased in the OLD group (4.8 mL/day) compared to that in the Y group (3.0 mL/day) but decreased in the YM (1.7 mL/day) and PM (1.5 mL/day) groups, without statistical significance (Fig. 3E). Urine osmolality was also increased in the OLD group (1176.7 mOsm/kg H2O) compared to that in the Y group (846.7 mOsm/kg H2O) but decreased in the YM (1098.3 mOsm/kg H2O) and PM (970.0 mOsm/kg H2O) groups, without statistical significance (Fig. 3F). Urinary excretion of Na+ was increased in the OLD group (336.7 mL/day) compared to that in the Y group (145.1 mL/day) but significantly decreased in the PM (60.0 mL/day, p < 0.05) and YM (91.0 mL/day) groups (Fig. 3G). These results suggest that YM and PM restore the kidney aging indicators in mice, despite no significant differences in the survival rates.
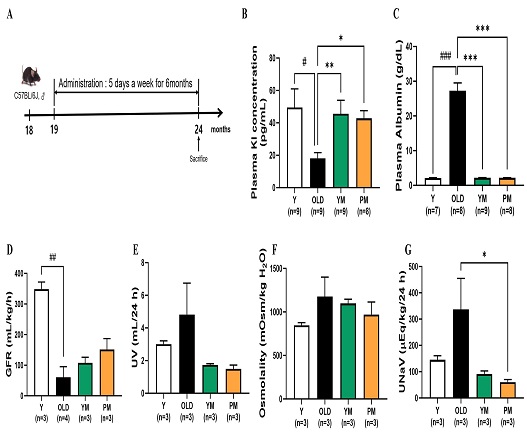
Effects of YM and PM on kidney aging indicators in mice. (A) Timeline for YM and PM administration in mice. Plasma levels of (B) klotho (Kl) and (C) albumin. (D) Glomerular filtration rate (GFR). (E) Urine volume (UV). (F) Urinary osmolality. (G) Urinary excretion of Na+ (UNaV). Values are represented as the mean ± standard error of the mean. *p < 0.05, **p < 0.01, and ***p < 0.001 determined via one-way analysis of variance (ANOVA), followed by Tukey’s multiple-comparison test. #p < 0.05, ##p < 0.01, and ###p < 0.001 determined via unpaired t-test between young and old groups.
4. YM and PM differentially regulate the kidney transcriptomics in aged mice
To investigate the mechanisms underlying these changes, RNA sequencing was performed on kidney tissues. Principal component analysis (PCA) was conducted to visualize gene expression data of the four groups (Y, OLD, YM, and PM). The resulting 3D PCA plot revealed clear clustering and separation among the groups (Fig. 4A). When the data were re-plotted, excluding the Y group data, the separation among the OLD, YM, and PM groups was even more distinct (Fig. 4B), indicating distinct gene expression patterns among all groups.
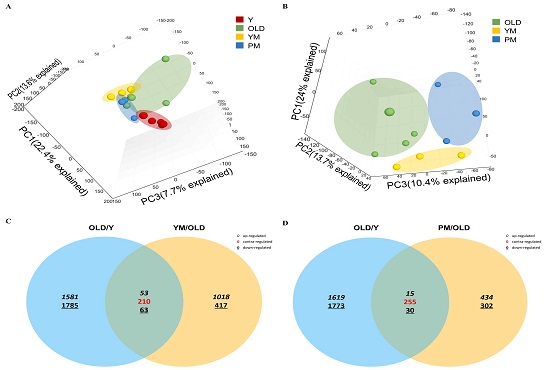
Comprehensive analyses of gene expression and differentially expressed genes (DEGs) in different age and treatment groups. (A) 3D principal component analysis plots illustrate the gene expression profiles of four groups: young (Y, red), old (OLD, green), old treated with YM (YM, yellow), and old treated with PM (PM, blue) groups. (B) Second plot only shows the OLD, YM, and PM groups, but not the Y group. Venn diagrams showing the overlapping DEGs between (C) OLD-Y and YM-OLD groups and (D) OLD-Y and PM-OLD groups.
To identify the genes driving these distinct clusters, we analyzed the DEGs in OLD-Y, YM-OLD, and PM-OLD comparisons and identified the contra-regulated genes (Fig. 4C and D). Specifically, 210 genes were contra-regulated between the OLD-Y and YM-OLD groups (Table S1), and 255 genes were contra-regulated between the OLD-Y and PM-OLD groups (Table S2). These genes possibly play critical roles in modulating the biomarkers associated with kidney aging.
5. Comparison of Gene Ontology (GO) and pathways regulated by YM and PM using kidney RNA sequencing data of aged mice
We conducted enrichment analysis of the differentially regulated genes in OLD-Y and YM-OLD (Fig. 5A). For genes downregulated in OLD-Y and upregulated in YM-OLD, significant GO terms included mitochondria, respiratory chain, and mitochondrial inner membrane. For genes upregulated in OLD-Y and downregulated in YM-OLD, significant GO terms included chromatin remodeling, nucleus, nucleoplasm, metal ion binding, transferase activity, kinase activity, protein serine/threonine kinase activity, and protein binding.
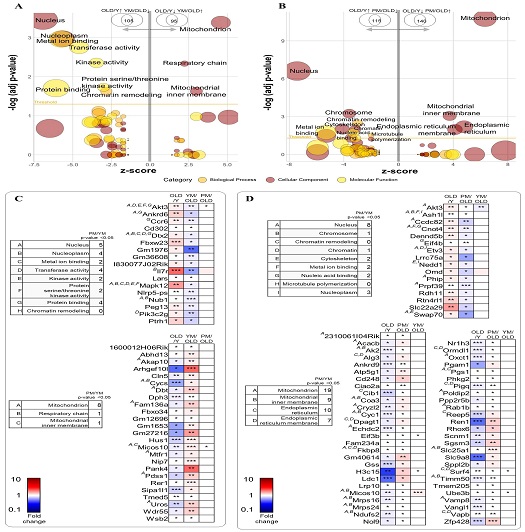
Gene expression and enrichment analyses of different age and treatment groups. (A and B) Bubble plots from Gene Ontology (GO) enrichment analysis obtained using the GO plot package in R based on Z-scores. (A) Left (negative Z-score): GO terms of 105 genes upregulated in the OLD-Y group and downregulated in the YM-OLD group. Right (positive Z-score): GO terms of 95 genes downregulated in the OLD-Y group and upregulated in the YM-OLD group. (B) Left: GO terms of 115 genes upregulated in the OLD-Y group and downregulated in the PM-OLD group. Right: GO terms of 140 genes downregulated in the OLD-Y group and upregulated in the PM-OLD group. (C and D) Heatmaps of genes showing opposite trends in OLD-Y vs. YM-OLD (C) and PM-OLD (D) groups, indicating the differentially regulated genes by YM and PM.
Similarly, we conducted enrichment analysis of genes differentially regulated in OLD-Y and PM-OLD (Fig. 5B). For genes downregulated in OLD-Y and upregulated in PM-OLD, significant GO terms included mitochondria, mitochondrial inner membrane, endoplasmic reticulum, and chromatin remodeling. For genes upregulated in OLD-Y and downregulated in YM-OLD, significant GO terms included chromatin remodeling, microtubule polymerization, nucleus, chromosome, chromatin, cytoskeleton, metal ion binding, and nucleic acid binding.
Comparative analysis revealed many shared GO terms between YM and PM due to the six common herbs. However, YM and PM exerted distinct clinical effects as PM contained two additional herbs that were not present in YM. To investigate these differences, we plotted a heatmap of the DEGs in YM and PM groups (Fig. 5C and D).
Although YM and PM regulated the same pathways, PM regulated 4.5-times more genes associated with mitochondria, mitochondrial inner membrane, endoplasmic reticulum, and endoplasmic reticulum membranes than YM. Moreover, YM, unlike PM, influenced transferase, kinase, protein serine/threonine kinase, and protein-binding activities. Conversely, PM, unlike YM, affected chromosomes, chromatin, cytoskeleton, nucleic acid binding, and microtubule polymerization. Both YM and PM affected the nucleus, nucleoplasm, metal ion binding, and chromatin remodeling. However, although both influenced some common genes in these terms, they also affected specific genes within shared categories.
6. YM and PM modulate the expression levels of hormone metabolism genes associated with longevity in aged mice
To investigate the hormonal mechanisms by which YM and PM affect aging, we conducted enrichment analysis focusing on genes involved in the regulation of hormone levels (GO:0010817; Table S3) with significant expression differences between the OLD and YM or PM groups.
In the YM-OLD comparison (Table S4), several key pathways were identified, including the longevity-regulating pathway (KEGG: 04211) and vasopressin-regulated water reabsorption pathway (KEGG: 04962). Key genes in these pathways were adiponectin receptor 1 (Adipor1), insulin receptor substrate 1 (Irs1), Kl, peroxisome proliferator-activated receptor gamma (Pparg), and protein kinase cAMP-activated catalytic subunit alpha (Prkaca) for longevity and dynein light chain LC8-type 1 (Dynll1), Prkaca, and syntaxin 4A (Stx4a) for vasopressin (Fig. 6A).
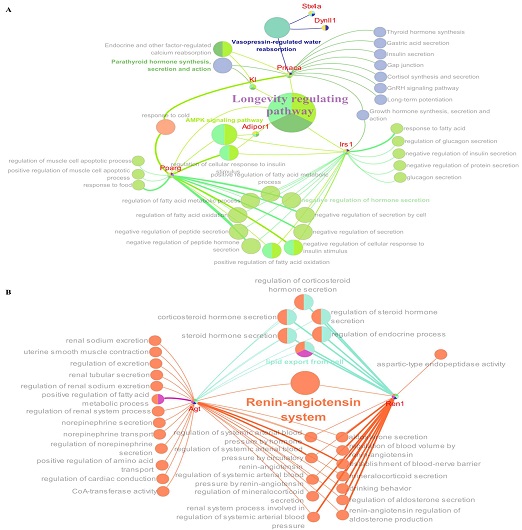
Network analysis of the biological pathways influenced by YM and PM based on enrichment analysis of hormone-related genes. (A) Network plot showing the pathways influenced by YM, including the longevity-regulating and vasopressin-regulated water reabsorption pathways. Key genes: adiponectin receptor 1 (Adipor1), insulin receptor substrate 1 (Irs1), klotho (KL), peroxisome proliferator-activated receptor gamma (Pparg), and protein kinase cAMP-activated catalytic subunit alpha (Prkaca) in the longevity pathway; syntaxin 4A (Stx4a), dynein light chain LC8-type 1 (Dynll1), and Prkaca in the vasopressin pathway; Prkaca in both pathways. (B) Network plot showing the renin–angiotensin system pathway influenced by PM. Key genes: angiotensinogen (Agt) and renin 1 (Ren1), highlighting connections to various hormonal mechanisms.
In the PM-OLD comparison (Table S5), the renin–angiotensin system (KEGG: 04614) was the key pathway, which is consistent with the historical use of PM. Key GO terms included aldosterone secretion (GO: 0035932), mineralocorticoid secretion (GO: 0035931), regulation of blood volume by renin–angiotensin (GO: 0002016), and regulation of systemic arterial blood pressure by circulatory renin–angiotensin (GO: 0001991), along with the important genes, angiotensinogen (Agt) and renin 1 (Ren1; Fig. 6B).
Discussion
We found that both YM and PM significantly increased mean lifespan of C. elegans. In a previous study12), YM at 2 mg/ml showed the highest lifespan-extending effect (46.19 %), which is similar to our results. Furthermore, we also found that at a concentration of 200 μg·mL−1, PM's lifespan-extending effect was 7.14 percentage points higher than that of YM, a finding not reported in previous literature. We observed that treating CC and AL individually resulted in an increased lifespan at lower concentrations. This suggests that the greater lifespan-extending effect of PM compared to that of YM may be attributed to these two herbs.
In aging mice, YM and PM significantly increased the levels of plasma Kl, which is an aging marker that plays a protective role in kidney function by inhibiting fibrosis and enhancing the ability of the kidneys to filter blood28,29). We also confirmed the improvement in kidney function in both YM- and PM-treated aged mice. Elevated plasma albumin levels in the OLD group were reduced by both YM and PM. Moreover, decreased GFR in the OLD group was increased after YM and PM treatments30). In contrast, increased urine volume in the OLD group was decreased by YM and PM. Despite the increase in urine osmolality, urine volume did not decrease in the OLD group, similar to that in the YM and PM groups, indicating that the kidneys in the OLD group exhibited impaired concentrating ability due to inefficient water reabsorption, leading to higher concentrations of solutes in the urine31). Urine sodium levels were higher in the OLD group than in the Y group but significantly decreased in the PM group (p < 0.05).
Relationship between urinary sodium levels and osmolality suggests that higher sodium excretion is associated with increased osmolality32). However, the significant reduction in urine sodium levels in the treatment groups indicated an improvement in the ability of the kidney to manage electrolyte balance, which is crucial for maintaining overall kidney function. Collectively, these findings suggested that YM and PM administration can ameliorate kidney aging in mice, despite the lack of significant differences in survival rates.
Enrichment analyses of DEGs revealed that YM and PM differentially regulated several biological processes and pathways. For YM, significant enrichment of GO terms in the cellular component category included mitochondria, respiratory chain, and mitochondrial inner membrane. Chromatin remodeling was notably enriched in the biological process category. In contrast, PM significantly enriched terms related to the mitochondria, endoplasmic reticulum, endoplasmic reticulum membrane, chromatin remodeling, and microtubule polymerization.
Comparison of the enrichment analysis results of YM and PM revealed many common GO terms shared by both treatments. However, YM and PM showed distinct effects as PM contains two additional herbs that are not present in YM. Notably, PM regulated 4.5-times more genes associated with the mitochondria and endoplasmic reticulum than YM. Notably, PM is traditionally used to support Kidney Yang, which in TKM refers to the body’s energy. The fact that PM has a much broader impact on mitochondrial-related than YM aligns with this historical context. Additionally, a recent study reported that AL affects kidney mitochondria33).
The kidneys have high energy demands, particularly for the reabsorption processes in the renal tubules. Thus, mitochondrial dysfunction can lead to impaired energy production, resulting in decreased kidney function, and contributing to kidney disease progression34). Mitochondrial biogenesis and functions are essential to protect against kidney damage and promote kidney repair35). Additionally, the mitochondrial respiratory chain is vital for ATP production, which is necessary for various cellular processes, including ion transport, maintenance of cellular homeostasis, and prevention of oxidative stress36). These reports suggest YM and PM ameliorate age-related kidney dysfunction.
Here, YM and PM regulated different pathways. YM primarily affected pathways related to enzyme activity, suggesting its roles in signal transduction and cellular metabolism. In contrast, PM influenced the cell structure and genetic regulation, indicating its roles in maintaining physical and genetic stability. Notably, both YM and PM affected the nucleus and nucleoplasm, altered the chromatin structure, and inhibited the binding of metal ions. Their common pathways are crucial for the core cellular functions. Therefore, YM and PM contribute to cellular health and longevity through distinct but complementary mechanisms.
Enrichment analysis of genes associated with hormone regulation revealed that YM significantly affected the vasopressin-regulated water reabsorption and longevity-regulating pathways. YM mainly regulated the genes, Adipor1, Irs1, Kl, Pparg, and Prkaca, which play crucial roles in metabolic processes, insulin signaling, aging, and cellular signaling. Although Prkaca is involved in both YM and PM pathways, YM plays a more important role in promoting longevity via vasopressin-regulated water reabsorption. Historically, YM has been used in TKM to treat fluid metabolism issues via vasopressin-regulated water reabsorption.
PM upregulated the genes involved in the renin–angiotensin system, Agt and Ren1. The renin–angiotensin system is vital for blood pressure regulation and fluid balance. Therefore, PM affects the kidney function and aging through this pathway. Enrichment analysis also revealed that PM significantly influenced the renin–angiotensin system, including pathways related to aldosterone and mineralocorticoid secretion, and regulation of systemic arterial blood pressure. These effects may be attributed to the inclusion of CC and AL in PM that reduced the angiotensin-converting enzyme activity.
This study suggests that, although YM and PM share common pathways, they have distinct hormonal mechanisms to exert anti-aging effects. YM enhances longevity and kidney function via metabolic and water reabsorption pathways, whereas PM primarily influences blood pressure and electrolyte balance through the renin–angiotensin system. In previous studies, the mTOR and AMPK pathways involved in metabolic regulation have demonstrated anti-aging effects37), and vasopressin and aquaporin pathways in the kidneys have shown impacts on aging38). The renin-angiotensin system has also been shown to influence aging by regulating angiotensin-converting enzymes inhibitors and angiotensin II receptor blockers39). Their distinct effects highlight the complex interplay among traditional herbal medicines and indicate the therapeutic potential of YM and PM. However, further research is necessary to elucidate the specific mechanisms by which these herbs affect longevity and kidney function, particularly via hormonal pathways.
One limitation of this study is that, while the pathways of YM and PM were presented through the results of gene set enrichment analysis, there is no concrete evidence to support these findings. Future research should focus on identifying biological markers targeting these pathways to provide a stronger basis for the conclusions. Secondly, although it is generally expected that albumin levels decrease with aging, this study found that the albumin concentrations in the control group and the YM and PM treatment groups were within the normal range40), while the OLD group exhibited abnormally high albumin levels. Although prior studies suggest that this increase may be related to dehydration41), this study could not specifically confirm whether such a phenomenon occurred. It is essential to verify this aspect in future studies to ensure reproducibility. Thirdly, as this study is an in vivo experimental study, it has limitations in claiming that YM and PM have lifespan extension effects or that the proposed pathways function as indicated. However, it can serve as a foundational reference for future clinical research, and I believe it could offer ideas for clinical analyses by simultaneously collecting data on body fluid content or blood pressure during YM and PM administration.
Conclusion
Overall, this study demonstrated the anti-aging effects of the traditional herbal medicines, YM and PM, and their potential to extend HLE by improving the kidney function and hormonal pathways. Our findings revealed the chemical compositions of YM and PM and demonstrated their potent lifespan-extending effects in a C. elegans model. RNA sequencing of aged mouse kidneys revealed that YM and PM modulated gene expression, reversed aging-related changes, and improved the functions associated with longevity. Our findings highlight YM and PM as natural agents to extend HLE with only a few side effects.
Acknowledgments
This study was supported by the Korea Institute of Oriental Medicine (KSN2312021) and a National Research Foundation of Korea (NRF) grant funded by the Korean government (NRF-2022R1A2C2092786).
Data Availability Statement
Supplementary tables are available at Zenodo (https://doi.org/10.5281/zenodo.14004658). RNA-seq data have been deposited into the National Center for Biotechnology Information Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE273090. All analysis results are reported in this article.
References
-
Guo J, Huang X, Dou L, Yan M, Shen T, Tang W, et al. Aging and aging-related diseases: from molecular mechanisms to interventions and treatments. Signal Transduct Target Ther 2022;7(1):391. https://pubmed.ncbi.nlm.nih.gov/36522308/, .
[https://doi.org/10.1038/s41392-022-01251-0]

-
Poganik JR, Gladyshev VN. We need to shift the focus of aging research to aging itself. Proc Natl Acad Sci U S A 2023;120(37):e2307449120. https://pubmed.ncbi.nlm.nih.gov/37682890/, .
[https://doi.org/10.1073/pnas.2307449120]

-
Partridge L, Fuentealba M, Kennedy BK. The quest to slow ageing through drug discovery. Nat Rev Drug Discov 2020;19(8):513-32. https://pubmed.ncbi.nlm.nih.gov/32467649/, .
[https://doi.org/10.1038/s41573-020-0067-7]

- GBD 2021 Demographics Collaborators. Demographics collaborators, 2024. Global age-sex-specific mortality, life expectancy, and population estimates in 204 countries and territories and 811 subnational locations, 1950–2021, and the impact of the COVID-19 pandemic: a comprehensive demographic analysis for the Global Burden of Disease Study 2021. Lancet 2021;403(10440):1989-2056. https://pubmed.ncbi.nlm.nih.gov/38484753/, .
-
Tsuji I.. Epidemiologic Research on Healthy Life Expectancy and Proposal for Its Extension: A, rev. English version of Japanese in the Journal of the Japan Medical Association 2019;148(9):1781-4. JMA J 2020;3(3):149-53. https://pubmed.ncbi.nlm.nih.gov/33150248/, .
[https://doi.org/10.31662/jmaj.2020-0027]

-
Chudasama YV, Khunti K, Gillies CL, Dhalwani NN, Davies MJ, Yates T, et al. Healthy lifestyle and life expectancy in people with multimorbidity in the UK Biobank: A longitudinal cohort study. PLoS Med 2020;17(9):e1003332. https://pubmed.ncbi.nlm.nih.gov/32960883/, .
[https://doi.org/10.1371/journal.pmed.1003332]

-
Fadnes LT, Økland JM, Haaland ØA, Johansson KA. Estimating impact of food choices on life expectancy: A modeling study. PLoS Med 2022;19(2):e1003889. https://pubmed.ncbi.nlm.nih.gov/35134067/, .
[https://doi.org/10.1371/journal.pmed.1003889]

-
Mohammed I, Hollenberg MD, Ding H, Triggle CR. A critical review of the evidence that metformin is a putative anti-aging drug that enhances Healthspan and extends lifespan. Front Endocrinol 2021;12:718942. https://pubmed.ncbi.nlm.nih.gov/34421827/, .
[https://doi.org/10.3389/fendo.2021.718942]

-
de Jager J, Kooy A, Lehert P, Wulffelé MG, van der Kolk J, Bets D, et al. Long term treatment with metformin in patients with type 2 diabetes and risk of vitamin B-12 deficiency: randomised placebo controlled trial. BMJ 2010;340:c2181. https://pubmed.ncbi.nlm.nih.gov/20488910/, .
[https://doi.org/10.1136/bmj.c2181]

-
Stevenson-Hoare J, Leonenko G, Escott-Price V. Comparison of long-term effects of metformin on longevity between people with type 2 diabetes and matched non-diabetic controls. BMC Public Health 2023;23(1):804. https://pubmed.ncbi.nlm.nih.gov/37131166/, .
[https://doi.org/10.1186/s12889-023-15764-y]

-
Kwon S, Kim YC, Park JY, Lee J, An JN, Kim CT, et al. The long-term effects of metformin on patients with Type 2 diabetic kidney disease. Diabetes Care 2020;43(5):948-55. https://pubmed.ncbi.nlm.nih.gov/32132005/, .
[https://doi.org/10.2337/dc19-0936]

-
Chen W, Wang J, Shi J, Yang X, Yang P, Wang N, et al. Longevity effect of Liuwei dihuang in both Caenorhabditis Elegans and Aged Mice. Aging Dis 2019;10(3):578-91. https://pubmed.ncbi.nlm.nih.gov/31165002/, .
[https://doi.org/10.14336/AD.2018.0604]

-
Sangha JS, Sun X, Wally OS, Zhang K, Ji X, Wang Z, et al. Liuwei Dihuang (LWDH), a traditional Chinese medicinal formula, protects against β-amyloid toxicity in transgenic Caenorhabditis elegans. PLoS ONE 2012;7(8):e43990. https://pubmed.ncbi.nlm.nih.gov/22952840/, .
[https://doi.org/10.1371/journal.pone.0043990]

-
Tseng YT, Chang FR, Lo YC. The Chinese herbal formula Liuwei dihuang protects dopaminergic neurons against Parkinson’s toxin through enhancing antioxidative defense and preventing apoptotic death. Phytomedicine 2014;21(5):724-33. https://pubmed.ncbi.nlm.nih.gov/24411708/, .
[https://doi.org/10.1016/j.phymed.2013.11.001]

- Hwang J, Jung H, Song B. Case report for kidney Yang deficiency syndrome Patient. The Journal of Internal Korean Medicine, 2014;35:349-53.
-
Isobe H, Yamamoto K, Cyong JC. Effects of hachimi-jio-gan (ba-wei-di-huang-wan) on blood flow in the human central retinal artery. Am J Chin Med 2003;31(3):425-35. https://pubmed.ncbi.nlm.nih.gov/12943173/, .
[https://doi.org/10.1142/S0192415X03001181]

- Jung H, Lee H, Lee J, Kim D. Clinical study of effect to the health-growth after the administration of Boyangsungjangtang to the prepuberty children. J Pediatr Korean Med 2001;15(1):47-57.
- Lee M, Son I. Effects of aqua-acupuncture of Yukmijihwangtang and Palmijihwangtang water extracts on the renal function. J Korean Acupunct Moxibustion Soc 1998;15(2):255-77.
- Seo Y, Lee E, Bae H, Shin M, Hong C. Antioxidant effects of the herbs composing Yukmijihwang-tang on PC12 cell. Korean J Orient Physiol Pathol 2003;17(1):203-8.
-
Neild GH. Life expectancy with chronic kidney disease: an educational review. Pediatr Nephrol 2017;32(2):243-8. https://pubmed.ncbi.nlm.nih.gov/27115888/, .
[https://doi.org/10.1007/s00467-016-3383-8]

-
Kim HS, Jeon DH, Oh MS. Healing effect of Yukmijihwang-tang on fracture factor and morphological changes in femur fractured mice. J Korean Med Rehabil 2020;30(4):17-30.
[https://doi.org/10.18325/jkmr.2020.30.4.17]

- Lee B. Symptom Identification Treatment. Seongbosa: Seoul, 1992, 467-8.
-
Roh M, Lee J, Jun H, Han Y, Kim H, Leem J. A Patient with Moderate Insomnia Improved after Treatment with Yukmijihwang-tang (Liuwei Dihuang tang), Hwangryunhaedok-tang (Huanglian Jiedu tang), and electroacupuncture: A Case Report. J Int Korean Med 2020;41(3):494-501.
[https://doi.org/10.22246/jikm.2020.41.3.494]

-
Park HH, Jung Y, Lee SV. Survival assays using Caenorhabditis elegans. Mol Cells 2017;40(2):90-9. https://pubmed.ncbi.nlm.nih.gov/28241407/, .
[https://doi.org/10.14348/molcells.2017.0017]

-
Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974;77(1):71-94. https://pubmed.ncbi.nlm.nih.gov/4366476/, .
[https://doi.org/10.1093/genetics/77.1.71]

-
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15-21. https://pubmed.ncbi.nlm.nih.gov/23104886/, .
[https://doi.org/10.1093/bioinformatics/bts635]

-
Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31(2):166-9. https://pubmed.ncbi.nlm.nih.gov/25260700/, .
[https://doi.org/10.1093/bioinformatics/btu638]

-
Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997;390(6655):45-51. https://pubmed.ncbi.nlm.nih.gov/9363890/, .
[https://doi.org/10.1038/36285]

-
Saito Y, Yamagishi T, Nakamura T, Ohyama Y, Aizawa H, Suga T, et al. Klotho protein protects against endothelial dysfunction. Biochem Biophys Res Commun 1998;248(2):324-29. https://pubmed.ncbi.nlm.nih.gov/9675134/, .
[https://doi.org/10.1006/bbrc.1998.8943]

-
Stevens LA, Coresh J, Greene T, Levey AS. Assessing kidney function—measured and estimated glomerular filtration rate. N Engl J Med 2006;354(23):2473-83. https://pubmed.ncbi.nlm.nih.gov/16760447/, .
[https://doi.org/10.1056/NEJMra054415]

-
Sands JM, Layton HE. The physiology of urinary concentration: an update. Semin Nephrol 2009;29(3):178-95. https://pubmed.ncbi.nlm.nih.gov/19523568/, .
[https://doi.org/10.1016/j.semnephrol.2009.03.008]

-
Reddi AS. Interpretation of urine electrolytes and osmolality, in: Fluid, Electrolyte and Acid-Base Disorders. Springer, Cham, 2018.
[https://doi.org/10.1007/978-3-319-60167-0]

-
Wu Z, Qian J, Feng C, Chen Z, Gao X, Liu Y, et al. A review of Aconiti Lateralis Radix Praeparata (Fuzi) for kidney disease: phytochemistry, toxicology, herbal processing, and pharmacology. Front Pharmacol 2024;15:1427333. https://pubmed.ncbi.nlm.nih.gov/39021829/, .
[https://doi.org/10.3389/fphar.2024.1427333]

-
Fontecha-Barriuso M, Lopez-Diaz AM, Guerrero-Mauvecin J, Miguel V, Ramos AM, Sanchez-Niño MD, et al. Tubular mitochondrial dysfunction, oxidative stress, and progression of chronic kidney disease. Antioxidants (Basel) 2022;11(7):1356. https://pubmed.ncbi.nlm.nih.gov/35883847/, .
[https://doi.org/10.3390/antiox11071356]

-
Tang W, Wei Q. The metabolic pathway regulation in kidney injury and repair. Front Physiol 2024;14:1344271. https://pubmed.ncbi.nlm.nih.gov/38283280/, .
[https://doi.org/10.3389/fphys.2023.1344271]

-
Ho HJ, Shirakawa H. Oxidative stress and mitochondrial dysfunction in chronic kidney disease. Cells 2022;12(1):88. https://pubmed.ncbi.nlm.nih.gov/36611880/, .
[https://doi.org/10.3390/cells12010088]

-
Surugiu R, Iancu MA, Vintilescu ȘB, Stepan MD, Burdusel D, Genunche-Dumitrescu AV, et al. Molecular Mechanisms of Healthy Aging: The Role of Caloric Restriction, Intermittent Fasting, Mediterranean Diet, and Ketogenic Diet-A Scoping Review. Nutrients. 2024;16(17):2878. https://pubmed.ncbi.nlm.nih.gov/39275194/, .
[https://doi.org/10.3390/nu16172878]

-
Tamma G, Goswami N, Reichmuth J, De Santo NG, Valenti G. Aquaporins, vasopressin, and aging: current perspectives. Endocrinology. 2015;156(3):777-88. https://pubmed.ncbi.nlm.nih.gov/25514088/, .
[https://doi.org/10.1210/en.2014-1812]

-
Egan BM, Scharf A, Pohl F, Kornfeld K. Control of aging by the renin-angiotensin system: a review of C. elegans, Drosophila, and mammals. Front Pharmacol. 2022;13:938650. https://pubmed.ncbi.nlm.nih.gov/36188619/, .
[https://doi.org/10.3389/fphar.2022.938650]

- Otto GP, Rathkolb B, Oestereicher MA, Lengger CJ, Moerth C, Micklich K, et al. Clinical Chemistry Reference Intervals for C57BL/6J, C57BL/6N, and C3HeB/FeJ Mice (Mus musculus). J Am Assoc Lab Anim Sci. 2016;55(4):375-86. https://pubmed.ncbi.nlm.nih.gov/27423143/, .
-
Miller GD, Teramoto M, Smeal SJ, Cushman D, Eichner D. Assessing serum albumin concentration following exercise-induced fluid shifts in the context of the athlete biological passport. Drug Test Anal. 2019;11(6):782-91. https://pubmed.ncbi.nlm.nih.gov/30690899/, .
[https://doi.org/10.1002/dta.2571]